Abstract
Introduction Patients with diffuse large B-cell lymphoma (DLBCL) are generally treated in the frontline setting with a standard immunochemotherapy regimen (R-CHOP), but for the 30-40% of patients who develop relapsed disease (relapsed-refractory DLBCL, rrDLBCL), prognosis is generally poor. While some novel therapies for rrDLBCL have measurably improved patient outcomes, there remains a notable proportion of patients for whom treatments fail or are not viable. Under the expectation that mutations contributing to treatment resistance will be selected for, and thus be enriched upon relapse, we sought to comprehensively explore the genetic features of rrDLBCL in comparison to the general DLBCL patient population.
Methods We applied whole genome sequencing (WGS) and whole exome sequencing (WES) to 107 tumour tissue biopsies and/or liquid biopsies collected from patients at relapse following R-CHOP and, in some cases, one or more salvage therapies. We supplemented this with rrDLBCL cases from previous publications for a total of 155 cases with exome data. Following Illumina sequencing, read alignment, and quality control, single nucleotide variants and small insertions/deletions (cumulative simple somatic mutations, SSMs) were detected using a consensus of 3/4 variant callers (Strelka2, MuTect2, SAGE, and LoFreq). We subsequently performed ultra-low pass WGS (lpWGS) (0.3-0.5x coverage) on 67 rrDLBCL liquid biopsies for a total of 222 cases with copy number variant (CNV) information. CNVs were identified using ichorCNA (lpWGS), Sequenza (WES), and Battenberg (WGS), and regions recurrently perturbed were identified using GISTIC2. The frequency of SSMs and CNVs was compared to a diagnostic cohort comprised of WES from Schmitz et al. (2018 NEJM) and BC cases we previously subjected to WGS. The genetic subgroup of each case was assigned using LymphGen.
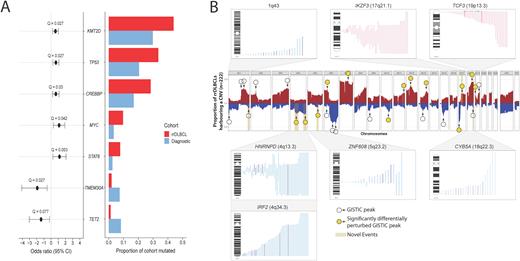
Results Comparing the landscape of SSMs between diagnostic and rrDLBCL revealed four genes mutated at significantly (Q < 0.1) higher rates among rrDLBCLs: KMT2D (40.7% of rrDLBCLs), TP53 (32.3%), STAT6 (7.1%), and MYC (9.03%). Three genes (TMEM30A, TET2, and BCL10) were significantly depleted for mutations at relapse (Figure 1A). Although our rrDLBCL cohort was generally balanced in terms of molecular subgroups (56 GCB, 52 ABC, 16 UNC), the prevalence of genetic subgroups varied, with EZB comprising more cases than any other class (26.3%), and a limited number classified as BN2 (7.5%), MCD (8.3%), ST2 (3.8%) or A53 (6.0%). Although MYC mutations, indicative of a MYC translocation, were enriched in rrDLBCL, we did not observe an over-representation of EZB-MYC+ cases. 42.1% of cases were unclassified via LymphGen, including 54.5% of ABC-rrDLBCLs.
Large arm-level or whole chromosome gains were common, particularly those involving chromosome 7 (47.7% of rrDLBCLs) or encompassing 18q23 (including BCL2, 44.1%), as were deletions affecting 6q23.3 (TNFAIP3, 45.0%) and 17p13 (TP53, 37.4%). We observed recurrent CNVs perturbing established DLBCL genes, including gains of REL and BCL2, and deletions of CDKN2A/CDKN2B, RB1, and PTEN. Comparing the frequency of recurrent CNVs to our diagnostic cohort revealed 14 regions significantly enriched for CNVs at relapse (Figure 1B). These included deletions of TP53, PTEN, and gains involving STAT6, MIR17HG and BCL2. We also noted several recurrent events that appeared exclusively in rrDLBCL that have not been previously described. These include recurrent deletions affecting the MHC Class I regulator IRF2 (4q34.3, 21.6% of rrDLBCLs) and the RNA splicing regulator HNRNPD (4q13.2, 22.5%) and gains perturbing the B-cell maturation factor IKZF3/Aiolos (17q21.1, 20.7%), and B-cell differentiation regulator TCF3 (19p13.3, 12.6%), with the latter event predominating in ABC-rrDLBCL.
Conclusions This cohort of rrDLBCL contained fewer BN2 cases and more EZB, which may relate in part to an enrichment of transformations from FL. The prevalence of fewer BN2 cases at relapse is consistent with the notion that these cases have superior outcomes afforded by R-CHOP. The depletion of mutations in TET2 and a low rate of ST2 cases could imply that such cases are also less likely to relapse on standard therapy. The frequent observation of deletions affecting HNRNPD points to an under-appreciated role of RNA-binding proteins in DLBCL relapse. Recurrent deletions of IRF2 could further explain immune evasion observed in rrDLBCL.
Disclosures
Rushton:SAGA Diagnostics: Consultancy. Alcaide:SAGA Diagnostics: Current Employment. Coyle:Allakos, Inc.: Consultancy; Dren Bio, Inc.: Consultancy. Michaud:Epizyme: Current Employment. Daigle:Epizyme: Current Employment. Hay:Roche: Research Funding; Karyopharm: Research Funding; AbbVie: Research Funding; Merck: Research Funding; Seagen: Research Funding. Jain:Incyte: Research Funding; MyeloidTx: Consultancy; BMS: Consultancy; Novartis: Consultancy; Kite Pharma: Consultancy, Research Funding. Kuruvilla:Gilead: Consultancy, Honoraria; Amgen: Honoraria; Medison Ventures: Consultancy; Antengene: Consultancy; Novartis: Honoraria; Janssen: Honoraria; Astra Zeneca: Honoraria, Research Funding; Roche: Consultancy, Honoraria, Research Funding; Merck: Consultancy, Honoraria, Research Funding; Karyopharm: Consultancy, Honoraria, Other: DSMB; Bristol Myers Squibb: Consultancy, Honoraria; Abbvie: Consultancy, Honoraria; Inctye: Honoraria; Seattle Genetics: Consultancy, Honoraria; Pfizer: Honoraria; Lymphoma Canada: Membership on an entity's Board of Directors or advisory committees. Crump:Roche: Research Funding; Kyte-Gilead, Novartis: Honoraria. Assouline:Genentech/Roche, Astra Zeneca, Novartis, BMS, Jazz, Gilead, Amgen, Beigene, Abbvie, Paladin: Consultancy, Honoraria; Novartis: Research Funding. Steidl:Abbvie: Consultancy; Bayer: Consultancy; Bristol Myers Squibb: Consultancy; Curis Inc: Consultancy; Epizyme: Research Funding; Roche: Consultancy; Seattle Genetics: Consultancy; Trillium Therapeutics: Research Funding. Scott:Janssen: Consultancy, Research Funding; NanoString: Patents & Royalties; Roche: Research Funding; Incyte: Consultancy; AstraZeneca: Consultancy, Honoraria; Abbvie: Consultancy. Johnson:Merck, AbbVie, Roche, Gilead: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal